Feature Sources
GAP can employ any type of biological data sources for extracting gene/protein features. For the current implementation, we have extracted seven features from two major sources of scientific texts, and online biological databases.
GAP adopts some external and well-developed semantic search engines (FACTA and GoPubMed) to annotate the potential interacting genes/proteins out of the PubMed documents. GAP, therefore, benefits from the state-of-the art natural language processing techniques to acquire a more comprehensive and accurate information about the genes of the interest.
Information on the selected features
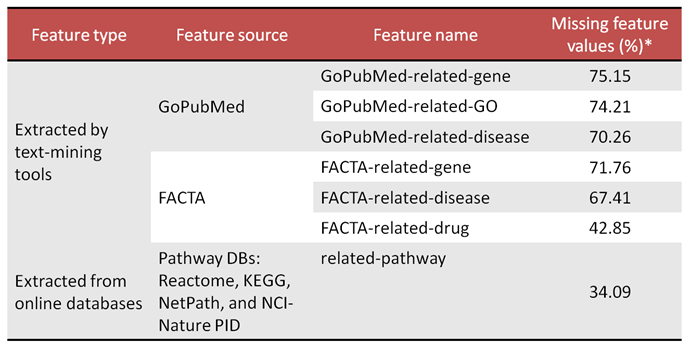
* Last column shows the percentage of the protein-coding genes for which the corresponding feature sets are empty according to the current version of the feature data sources
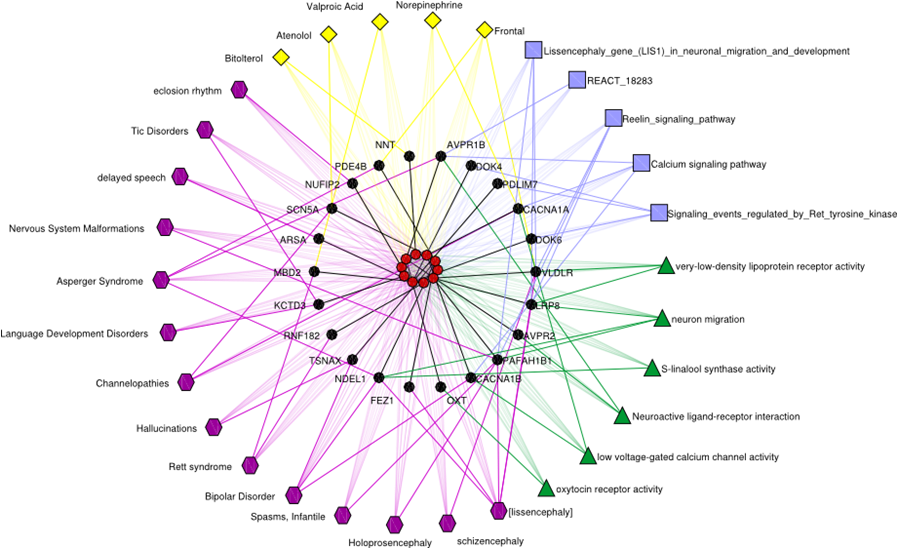
Figure: Sample of GAP prediction on autism-related genes and evidences from the related diseases, drugs, GO terms, and pathways.