Niu, Y., Otasek, D., Jurisica, I. Evaluation of linguistic features useful in extraction of interactions from PubMed; Application to annotating known, high-throughput and predicted interactions in I2D. Bioinformatics, 2009. doi: 10.1093/bioinformatics/btp602.
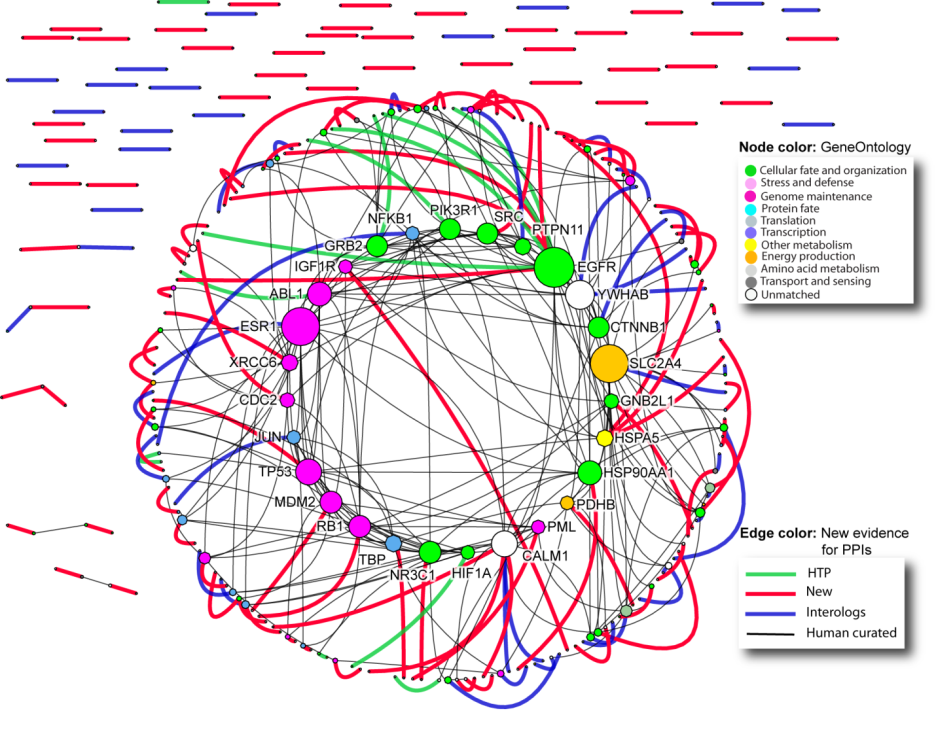
Protein-protein interactions with new evidence from
automatic text mining. Expanding the original set of 1,433 proteins and 845 interactions
by including interactions from I2D results in a network with 8,250 proteins and 21,652
interactions. Subgraph from Supplementary Figure 3, showing only the largest
connected component and highlighting new evidence, new evidence for human curated
PPIs, predicted PPIs and PPIs from high-throughput experiments. Combined network
comprises 392 interactions on 322 proteins. Node color is according to GeneOntology,
as shown in the legend. Node size is based on node degree in the network. For
simplicity, only protein names of high degree nodes are shown. Edge color is according
to the PPI source, as described in the legend.